COVID-19 vs the Spanish flu: Compare with caution
Why has COVID-19 defied the patterns followed by influenza?

Just over 100 years ago, as the world was emerging from World War I, a silent killer started making its way from the trenches. Unknowingly, soldiers returning home carried the Spanish influenza to busy railway stations, big cities, quiet towns and rural areas. What followed was the 1918-19 flu pandemic, which infected about 500 million people – a third of the world’s population then. Fifty million people died.
As COVID-19 took hold in 2020, people have been looking back on the 1918-19 influenza pandemic mainly for one reason: how and when is the current pandemic going to end? But despite some similarities between the two viruses, the Spanish flu pandemic is not a reliable indicator of what course COVID-19 will take.
What COVID and influenza have in common
COVID-19 and the Spanish flu are airborne diseases with similar basic reproductive number. The so-called R, which shows how many people someone will infect is between 2 and 4 for both viruses.
SARS-CoV-2 and H1N1 are also very transmissible in super-spreader events, such as concerts, football matches and religious worship.
Crucially, both viruses contain hemagglutinin – the protein which allows them to replicate through receptor binding and membrane fusion with the human host’s cells.
Another common feature is the viruses’ ability to trigger a cytokine storm. It is an overreaction of the immune system, which happens when the body, in an attempt to defeat the virus, attacks itself instead, leading to fluid build-up and hyperinflammation of the lungs. This helps explain why young, healthy people fell severely ill with the Spanish flu and why we see similar cases happen due to COVID today, albeit at a much lower level.
Where the differences begin
The novel coronavirus and the 1918 influenza are structured differently, which affects their ability to mutate, as well as the frequency of the mutations.
Influenza’s genetic material is organised in segmented chunks, which enables its rapid evolution, as the virus can trade entire RNA segments with other influenza viruses. Its signature seasonality also contributes to its fast mutation, as usually several flu strains circulate during the winter period. However, it also means it is a lot less common in spring and summer, pretty much eliminating the possibility of a significant outbreak during this time of the year.
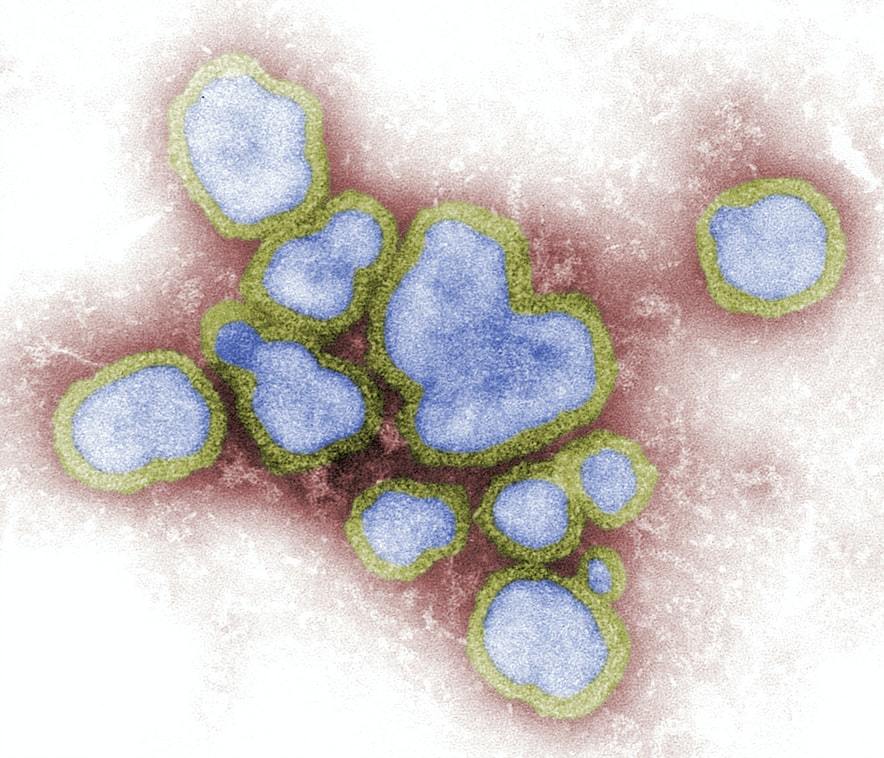
Unlike influenza and other RNA viruses, coronaviruses double-check copied genes and correct errors made during replication. This explains why only a handful of SARS-CoV-2 mutations have been identified so far, despite the virus travelling through multiple generations of human hosts on different continents.
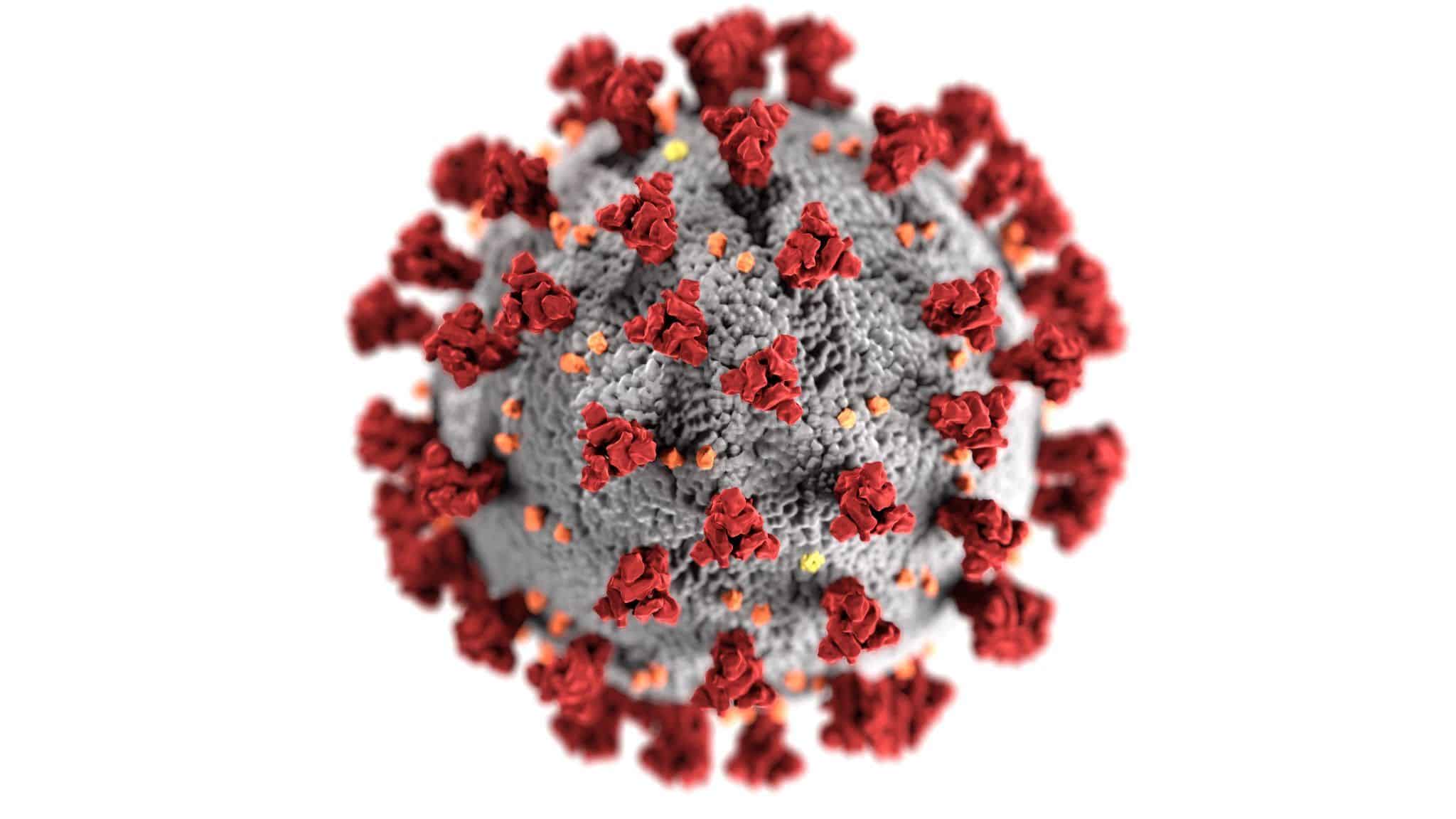
The novel coronavirus has also proved relatively stable in different climates. Any curb in the number of infections during the warmer months was the result of strict social-distancing measures, rather than the rise of temperatures.
This means that future peaks of the disease are less likely to be driven by mutations and change of seasons, making it harder to predict the next wave of infections or when it will come to an end. But, if it is changes in human behaviour that determine the progress of the pandemic, rather than changes in SARS-CoV-19 itself, we might have more control than we think.